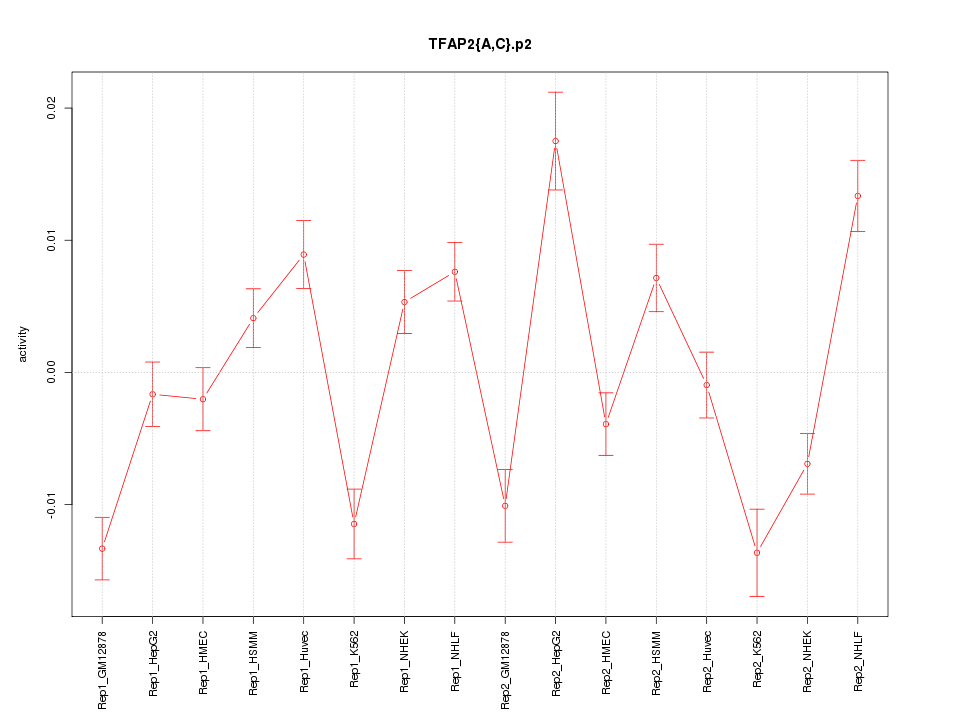
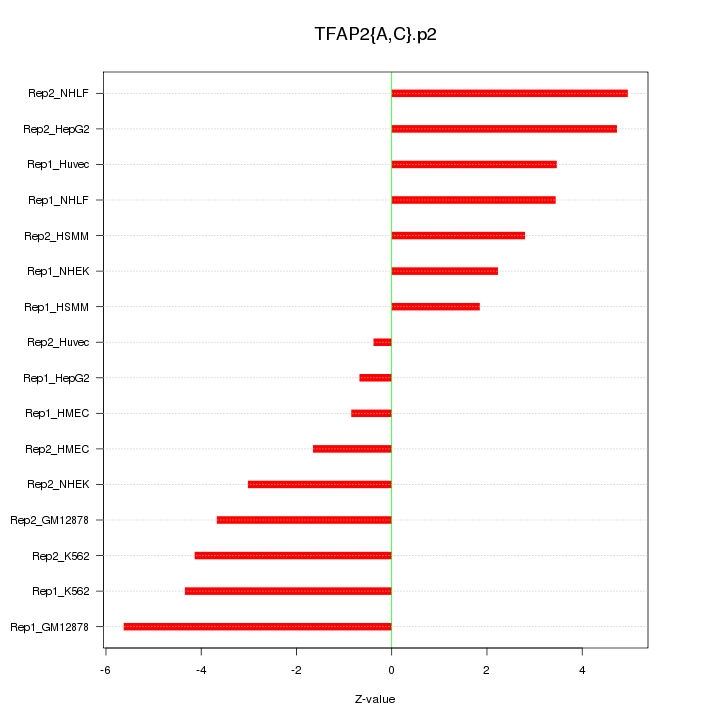
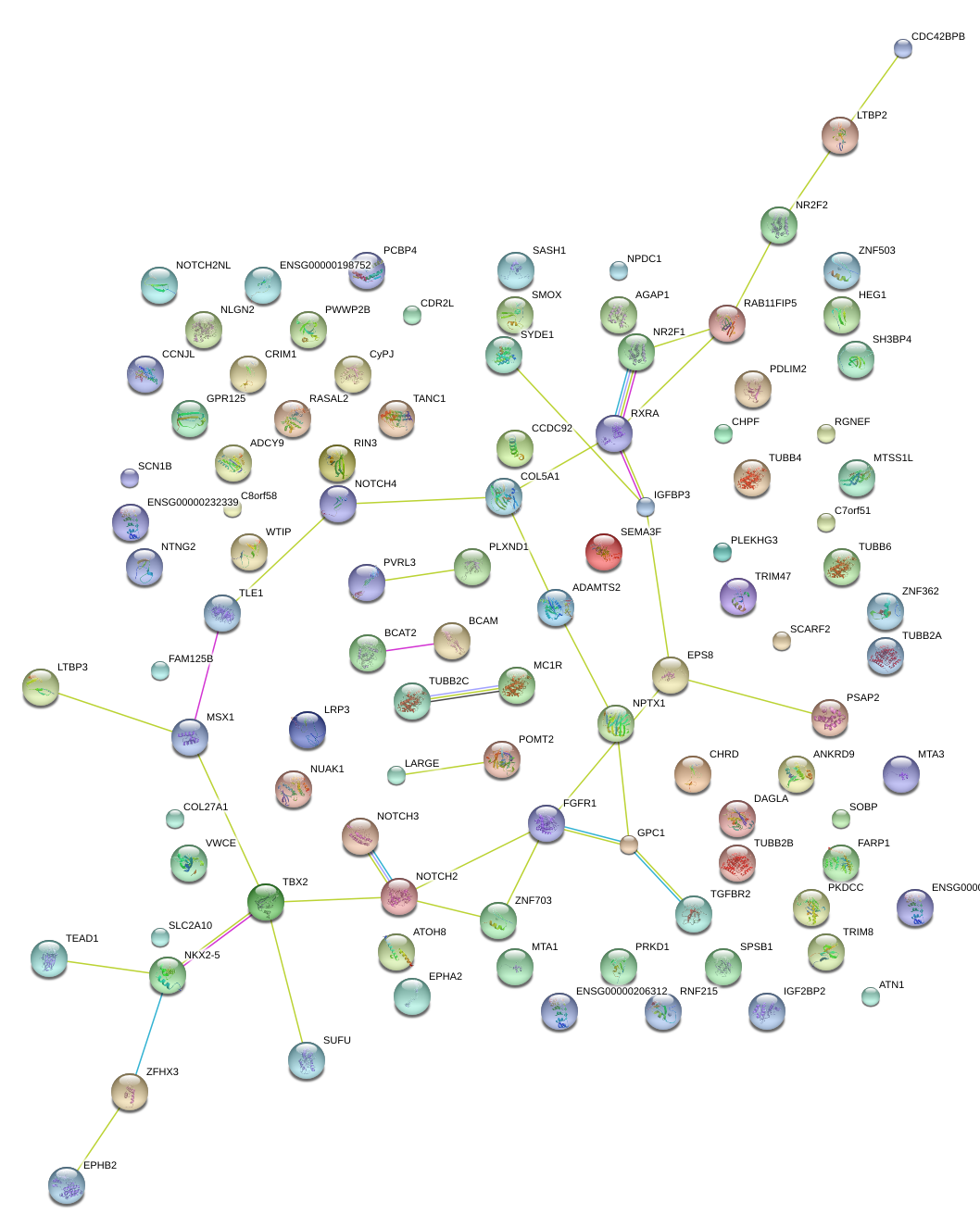
|
chr9_+_115958051
|
6.883
|
NM_032888
|
COL27A1
|
collagen, type XXVII, alpha 1
|
|
chr9_+_136673534
|
5.801
|
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr2_+_236068029
|
5.442
|
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr5_+_92946348
|
5.349
|
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr1_+_33494745
|
4.954
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr9_+_136673464
|
4.634
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr17_+_56832255
|
4.396
|
|
TBX2
|
T-box 2
|
|
chr3_-_126257425
|
4.183
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr3_-_130807865
|
4.058
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr17_+_56831951
|
3.984
|
NM_005994
|
TBX2
|
T-box 2
|
|
chr8_-_38444982
|
3.881
|
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr2_+_235525355
|
3.869
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr8_+_37672427
|
3.742
|
NM_025069
|
ZNF703
|
zinc finger protein 703
|
|
chr5_-_159672060
|
3.610
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chr2_+_236067687
|
3.585
|
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr3_-_187025501
|
3.412
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr19_+_15079141
|
3.307
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr19_+_38377329
|
3.290
|
NM_002333
|
LRP3
|
low density lipoprotein receptor-related protein 3
|
|
chr3_+_185580554
|
3.230
|
NM_003741
|
CHRD
|
chordin
|
|
chr22_+_49459935
|
3.209
|
NM_001080420
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3
|
|
chr20_+_4077466
|
3.196
|
|
SMOX
|
spermine oxidase
|
|
chr3_+_50217692
|
3.188
|
NM_006841
|
SLC38A3
|
solute carrier family 38, member 3
|
|
chr19_+_39664706
|
3.174
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr2_+_42128521
|
3.151
|
NM_138370
|
PKDCC
|
protein kinase domain containing, cytoplasmic homolog (mouse)
|
|
chr22_-_32646325
|
3.138
|
NM_004737
NM_133642
|
LARGE
|
like-glycosyltransferase
|
|
chr17_+_56832492
|
3.114
|
|
TBX2
|
T-box 2
|
|
chr19_+_40213431
|
3.091
|
NM_001037
NM_199037
|
SCN1B
|
sodium channel, voltage-gated, type I, beta
|
|
chr9_+_115957537
|
3.039
|
|
COL27A1
|
collagen, type XXVII, alpha 1
|
|
chr2_+_36436316
|
3.021
|
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr3_-_187025438
|
2.992
|
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr1_+_22909916
|
2.938
|
NM_004442
NM_017449
|
EPHB2
|
EPH receptor B2
|
|
chr19_+_50004134
|
2.906
|
NM_001013257
NM_005581
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group)
|
|
chr19_-_15172773
|
2.800
|
NM_000435
|
NOTCH3
|
notch 3
|
|
chr2_+_241023908
|
2.785
|
|
GPC1
|
glypican 1
|
|
chr10_+_104394185
|
2.749
|
NM_030912
|
TRIM8
|
tripartite motif containing 8
|
|
chr12_-_123023312
|
2.721
|
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr9_+_134026836
|
2.627
|
|
NTNG2
|
netrin G2
|
|
chr4_-_22126437
|
2.565
|
NM_145290
|
GPR125
|
G protein-coupled receptor 125
|
|
chr17_-_71385651
|
2.560
|
|
TRIM47
|
tripartite motif containing 47
|
|
chr2_+_85834111
|
2.550
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr20_+_44771532
|
2.522
|
NM_030777
|
SLC2A10
|
solute carrier family 2 (facilitated glucose transporter), member 10
|
|
chr9_-_83493415
|
2.503
|
NM_005077
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr11_+_61204392
|
2.359
|
NM_006133
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chr2_+_236067424
|
2.350
|
NM_001037131
NM_014914
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr11_-_60819110
|
2.341
|
|
VWCE
|
von Willebrand factor C and EGF domains
|
|
chr11_-_60819273
|
2.307
|
NM_152718
|
VWCE
|
von Willebrand factor C and EGF domains
|
|
chr22_-_19122107
|
2.295
|
NM_153334
NM_182895
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr14_-_29466562
|
2.238
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr11_-_65082208
|
2.233
|
NM_001130144
NM_001164266
NM_021070
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
|
chr8_-_38444675
|
2.215
|
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr17_+_70495318
|
2.204
|
NM_014603
|
CDR2L
|
cerebellar degeneration-related protein 2-like
|
|
chr17_-_71386229
|
2.197
|
NM_033452
|
TRIM47
|
tripartite motif containing 47
|
|
chr11_-_65081967
|
2.182
|
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
|
chr6_+_107917965
|
2.171
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr9_+_136358145
|
2.161
|
|
RXRA
|
retinoid X receptor, alpha
|
|
chr1_+_9275542
|
2.153
|
|
SPSB1
|
splA/ryanodine receptor domain and SOCS box containing 1
|
|
chr14_+_104957298
|
2.142
|
|
MTA1
|
metastasis associated 1
|
|
chr16_-_4106022
|
2.139
|
|
ADCY9
|
adenylate cyclase 9
|
|
chr1_+_9275518
|
2.123
|
NM_025106
|
SPSB1
|
splA/ryanodine receptor domain and SOCS box containing 1
|
|
chr16_-_4106186
|
2.110
|
NM_001116
|
ADCY9
|
adenylate cyclase 9
|
|
chr14_-_76856776
|
2.053
|
NM_013382
|
POMT2
|
protein-O-mannosyltransferase 2
|
|
chr2_-_73193566
|
2.049
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr22_-_29113301
|
2.047
|
NM_001017981
|
RNF215
|
ring finger protein 215
|
|
chr17_-_76064842
|
2.040
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr7_-_45927263
|
2.033
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr10_+_134060681
|
2.023
|
NM_001098637
NM_138499
|
PWWP2B
|
PWWP domain containing 2B
|
|
chr5_+_72957736
|
1.995
|
NM_001080479
NM_001177693
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr9_+_128128913
|
1.995
|
NM_001011703
NM_033446
|
FAM125B
|
family with sequence similarity 125, member B
|
|
chr3_+_112273464
|
1.992
|
NM_015480
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr15_+_94674849
|
1.984
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr14_+_92049396
|
1.978
|
NM_024832
|
RIN3
|
Ras and Rab interactor 3
|
|
chr12_+_6907646
|
1.974
|
NM_001940
|
ATN1
|
atrophin 1
|
|
chr11_+_12652427
|
1.972
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr16_+_88517187
|
1.969
|
NM_006086
|
TUBB3
|
tubulin, beta 3
|
|
chr2_-_220116508
|
1.967
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr5_-_172594691
|
1.964
|
NM_001166175
NM_001166176
NM_004387
|
NKX2-5
|
NK2 transcription factor related, locus 5 (Drosophila)
|
|
chr14_+_64240902
|
1.952
|
NM_015549
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr1_-_16355013
|
1.944
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr4_+_4912272
|
1.942
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chr14_-_102593264
|
1.937
|
|
|
|
|
chr5_-_178704934
|
1.934
|
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr14_+_64240996
|
1.930
|
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr13_+_97593711
|
1.929
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr2_-_73193581
|
1.923
|
NM_015470
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr12_-_105056577
|
1.899
|
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr9_-_139060285
|
1.884
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr2_+_241023703
|
1.873
|
NM_002081
|
GPC1
|
glypican 1
|
|
chr20_+_4077409
|
1.860
|
NM_175839
NM_175840
NM_175841
NM_175842
|
SMOX
|
spermine oxidase
|
|
chr2_+_159533329
|
1.858
|
NM_001145909
NM_033394
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1
|
|
chr3_-_51976428
|
1.839
|
NM_020418
NM_001174100
NM_033008
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr6_+_148705646
|
1.838
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr3_+_30623376
|
1.809
|
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr14_-_102045849
|
1.797
|
NM_152326
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr10_-_76829857
|
1.781
|
|
ZNF503
|
zinc finger protein 503
|
|
chr16_-_71639670
|
1.773
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr1_+_176330254
|
1.762
|
|
RASAL2
|
RAS protein activator like 2
|
|
chr2_-_73193419
|
1.761
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr2_+_42129440
|
1.760
|
|
|
|
|
chr10_+_104253716
|
1.751
|
|
SUFU
|
suppressor of fused homolog (Drosophila)
|
|
chr16_-_69277355
|
1.743
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr8_+_22492586
|
1.736
|
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr14_-_102593384
|
1.721
|
NM_006035
|
CDC42BPB
|
CDC42 binding protein kinase beta (DMPK-like)
|
|
chr3_+_50167851
|
1.718
|
NM_004186
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F
|
|
chr7_+_99919485
|
1.713
|
NM_173564
|
C7orf51
|
chromosome 7 open reading frame 51
|
|
chr22_-_34754343
|
1.709
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr17_+_7252225
|
1.694
|
NM_020795
|
NLGN2
|
neuroligin 2
|
|
chr12_-_15833617
|
1.693
|
NM_004447
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr1_-_120413542
|
1.688
|
|
NOTCH2
|
notch 2
|
|
chr9_-_135847106
|
1.686
|
NM_001134398
NM_003371
|
VAV2
|
vav 2 guanine nucleotide exchange factor
|
|
chr16_-_49742651
|
1.682
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr16_+_85158357
|
1.674
|
NM_005251
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1)
|
|
chr13_+_26029799
|
1.663
|
NM_006646
|
WASF3
|
WAS protein family, member 3
|
|
chr4_+_995609
|
1.663
|
NM_001004356
NM_001004358
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr7_-_130069399
|
1.658
|
NM_138693
|
KLF14
|
Kruppel-like factor 14
|
|
chr12_-_49897743
|
1.657
|
|
POU6F1
|
POU class 6 homeobox 1
|
|
chr19_+_7486987
|
1.652
|
NM_018083
|
ZNF358
|
zinc finger protein 358
|
|
chr2_-_128792515
|
1.651
|
NM_004807
|
HS6ST1
|
heparan sulfate 6-O-sulfotransferase 1
|
|
chr5_-_57791544
|
1.637
|
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr14_-_24588916
|
1.632
|
NM_014178
|
STXBP6
|
syntaxin binding protein 6 (amisyn)
|
|
chr9_-_38059082
|
1.628
|
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr10_+_124124196
|
1.612
|
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr2_+_36436873
|
1.611
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr12_-_105165805
|
1.602
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr6_+_35335278
|
1.602
|
|
ZNF76
|
zinc finger protein 76
|
|
chr2_-_96899326
|
1.602
|
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C
|
|
chr10_+_123912930
|
1.599
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr20_+_1822943
|
1.588
|
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr1_-_120413783
|
1.585
|
|
NOTCH2
|
notch 2
|
|
chr1_-_120413698
|
1.583
|
|
NOTCH2
|
notch 2
|
|
chr8_+_25098296
|
1.581
|
|
DOCK5
|
dedicator of cytokinesis 5
|
|
chr16_-_69277194
|
1.580
|
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr22_-_19122047
|
1.575
|
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr11_+_19691397
|
1.574
|
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr22_+_27609889
|
1.567
|
NM_032173
|
ZNRF3
|
zinc and ring finger 3
|
|
chr13_+_26029879
|
1.567
|
|
WASF3
|
WAS protein family, member 3
|
|
chr10_-_76831430
|
1.552
|
NM_032772
|
ZNF503
|
zinc finger protein 503
|
|
chr18_-_53621277
|
1.551
|
NM_005603
|
ATP8B1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1
|
|
chr3_-_129689394
|
1.545
|
NM_001145662
|
GATA2
|
GATA binding protein 2
|
|
chr16_-_52877467
|
1.544
|
|
IRX3
|
iroquois homeobox 3
|
|
chr19_-_54557450
|
1.540
|
NM_003598
|
TEAD2
|
TEA domain family member 2
|
|
chr2_+_173000590
|
1.538
|
|
ITGA6
|
integrin, alpha 6
|
|
chr16_-_71650034
|
1.534
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chrX_-_54538591
|
1.532
|
|
FGD1
|
FYVE, RhoGEF and PH domain containing 1
|
|
chr1_-_212791467
|
1.532
|
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr19_-_50963942
|
1.531
|
|
|
|
|
chr1_-_85703198
|
1.524
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr20_+_1823825
|
1.523
|
NM_001040023
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr10_-_76831142
|
1.522
|
|
ZNF503
|
zinc finger protein 503
|
|
chr6_+_151603346
|
1.521
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr20_-_49818268
|
1.511
|
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr7_+_55054416
|
1.507
|
|
EGFR
|
epidermal growth factor receptor
|
|
chr16_-_4105703
|
1.504
|
|
ADCY9
|
adenylate cyclase 9
|
|
chr11_+_119586879
|
1.501
|
NM_178507
|
OAF
|
OAF homolog (Drosophila)
|
|
chr7_-_27120015
|
1.497
|
|
HOXA3
|
homeobox A3
|
|
chr12_-_47468985
|
1.495
|
NM_020983
|
ADCY6
|
adenylate cyclase 6
|
|
chr1_+_19843261
|
1.495
|
NM_005380
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr14_+_54102518
|
1.489
|
|
LOC644925
|
hypothetical LOC644925
|
|
chr6_-_128883125
|
1.487
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr11_-_6633427
|
1.486
|
NM_003737
|
DCHS1
|
dachsous 1 (Drosophila)
|
|
chr9_-_139060377
|
1.486
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr14_-_102045749
|
1.481
|
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr19_+_49943773
|
1.477
|
NM_005178
|
BCL3
|
B-cell CLL/lymphoma 3
|
|
chr9_-_139060416
|
1.472
|
NM_015392
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr2_-_218551786
|
1.470
|
|
TNS1
|
tensin 1
|
|
chr6_-_132314004
|
1.468
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr9_-_138126917
|
1.467
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr10_-_15453060
|
1.465
|
NM_001010924
|
FAM171A1
|
family with sequence similarity 171, member A1
|
|
chr22_-_28972683
|
1.465
|
NM_002309
|
LIF
|
leukemia inhibitory factor (cholinergic differentiation factor)
|
|
chr1_-_120413798
|
1.454
|
NM_024408
|
NOTCH2
|
notch 2
|
|
chr5_-_16989371
|
1.451
|
NM_012334
|
MYO10
|
myosin X
|
|
chr6_+_5030718
|
1.437
|
NM_001145115
|
PPP1R3G
|
protein phosphatase 1, regulatory (inhibitor) subunit 3G
|
|
chr3_+_189354167
|
1.437
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr3_+_51403760
|
1.435
|
NM_013286
|
RBM15B
|
RNA binding motif protein 15B
|
|
chr7_+_3307445
|
1.434
|
NM_152744
|
SDK1
|
sidekick homolog 1, cell adhesion molecule (chicken)
|
|
chr8_-_18915443
|
1.431
|
NM_015310
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr3_+_148610516
|
1.430
|
|
ZIC1
|
Zic family member 1 (odd-paired homolog, Drosophila)
|
|
chr10_+_101079099
|
1.429
|
|
CNNM1
|
cyclin M1
|
|
chr2_+_231610438
|
1.426
|
NM_001144994
|
C2orf72
|
chromosome 2 open reading frame 72
|
|
chr15_+_64782468
|
1.425
|
|
SMAD6
|
SMAD family member 6
|
|
chr15_+_65145215
|
1.419
|
|
SMAD3
|
SMAD family member 3
|
|
chr10_-_99084254
|
1.411
|
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2
|
|
chr5_-_178705036
|
1.409
|
NM_014244
NM_021599
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr4_-_5945596
|
1.408
|
NM_001014809
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr20_+_11819364
|
1.391
|
NM_181443
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chrX_+_46963107
|
1.390
|
|
CDK16
|
cyclin-dependent kinase 16
|
|
chr6_+_157722467
|
1.389
|
NM_024630
NM_153746
|
ZDHHC14
|
zinc finger, DHHC-type containing 14
|
|
chr11_+_72697310
|
1.387
|
NM_014786
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17
|
|
chr18_-_33399601
|
1.384
|
|
CELF4
|
CUGBP, Elav-like family member 4
|
|
chr10_+_104253706
|
1.370
|
NM_001178133
NM_016169
|
SUFU
|
suppressor of fused homolog (Drosophila)
|
|
chr7_-_27136876
|
1.368
|
NM_002141
|
HOXA4
|
homeobox A4
|
|
chr5_+_14196287
|
1.358
|
NM_007118
|
TRIO
|
triple functional domain (PTPRF interacting)
|
|
chr20_+_2621193
|
1.353
|
NM_001110514
|
EBF4
|
early B-cell factor 4
|
|
chr10_-_118491971
|
1.350
|
NM_025015
|
HSPA12A
|
heat shock 70kDa protein 12A
|
|
chr10_+_59942874
|
1.346
|
NM_001080512
|
BICC1
|
bicaudal C homolog 1 (Drosophila)
|
|
chr7_-_5429702
|
1.342
|
NM_001080495
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr19_+_38979502
|
1.340
|
NM_001129994
NM_001129995
NM_024076
|
KCTD15
|
potassium channel tetramerisation domain containing 15
|
|
chr19_-_60320592
|
1.338
|
NM_017607
|
PPP1R12C
|
protein phosphatase 1, regulatory (inhibitor) subunit 12C
|
|
chr22_+_22445035
|
1.332
|
NM_005940
|
MMP11
|
matrix metallopeptidase 11 (stromelysin 3)
|
|
chr12_+_6363482
|
1.332
|
NM_002342
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3)
|
|
chr20_+_1822779
|
1.329
|
NM_001040022
|
SIRPA
|
signal-regulatory protein alpha
|